Extending sfftk
Todo
multifile parser
Adding A Segmentation File Format
There are five (5) steps involved.
Step I: Create a reader module
Step II: Create a format adapter module
Step III: Implement a convert hook in the
sfftk.sff.handle_convert()functionStep IV: Add test data files to the
sfftk/test_datafolderStep V: Add unit tests in the
sfftk.unittestsmodule
Step I: Create a reader module
The role of the reader module is to be the first point of contact with the application-specific segmentation file.
Each reader module must implement a get_data(fn, *args, **kwargs) function which takes the segmentation file’s name as a string together with optional positional and keyword arguments to be passed down the class chain. This function then returns a single ad hoc object that encapsulates the segmentation with a simple API faithful to the segmentation’s structure to access various attributes of the segmentation e.g. headers, segments etc.
The reader module must then be placed in the sfftk.readers package and should be named as the extension followed by the word ‘reader’ e.g. for Segger files with a .seg extension this would be segreader.py.
Step II: Create a format adapter module
The format adapter is where the ad hoc segmentation object is adapted to the EMDB-SFF model.
The classes defined in this module represent a hierarchy describing a typical segmentation data structure. For the following discussion, please refer to the diagram below. (You might need to right-click and select an option to view the image in a new tab to see the image details.)
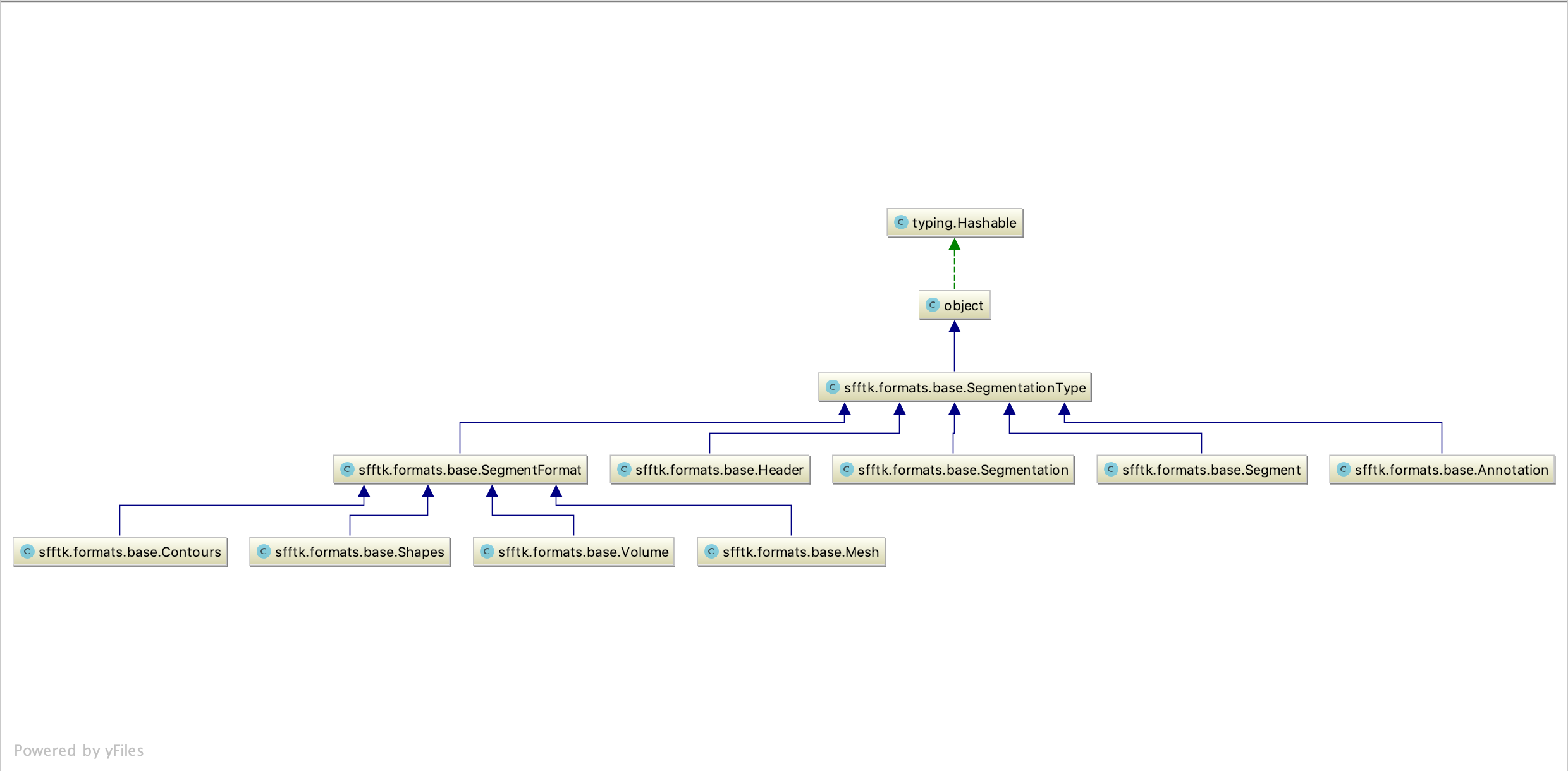
Note
Note that all classes are subclasses of sfftk.formats.base.SegmentationType, which may specify attributes and methods that apply to all subclasses. Currently, none are defined but it is envisaged that some may arise.
At the top of the hierarchy is a sfftk.formats.base.Segmentation subclass which hosts objects of two other classes:
a
sfftk.formats.base.Headersubclass for all header related metadata such as segment colours, names, segmentation space parameters such as the bounding box etc., andan iterable attribute called
segmentswhich is a container forsfftk.formats.base.Segmentsubclasses, each holding data for a single segment.
Each sfftk.formats.base.Segment subclass will in turn host objects of the following classes:
a
sfftk.formats.base.Annotationsubclass for segment annotation, andone or more segment format subclasses:
sfftk.formats.base.Volumefor 3D volumes,sfftk.formats.base.Meshfor meshes, and/orsfftk.formats.base.Shapesfor shape primitives
Format adapter modules must be placed in the sfftk.formats package and should be named as the segmentation file format’s extension e.g. for Segger files with a .seg extension this is seg.py.
Conversion is effected within each class by implementing a convert() method which takes the attributes obtained from the ad hoc segmentation object and uses them to populate fields in a corresponding EMDB-SFF attribute object. Given the hierarchy described above, the convert() method for objects higher up in the hierarchy may then invoke the convert() method of lower level objects when effecting their conversions. An example will be provided below.
Example:
To make the above concrete, we outline how the sfftk.formats.seg format adapter is implemented.
Segger segmentation files are represented using 3D volumes. Therefore, the sfftk.formats.seg module has the following top level structure:
from .base import Segmentation, Header, Segment, Annotation, Volume
import sfftkrw as schema
from ..readers import segreader
class SeggerAnnotation(Annotation):
pass
class SeggerVolume(Volume):
pass
class SeggerSegment(Segment):
pass
class SeggerHeader(Header):
pass
class SeggerSegmentation(Segmentation):
pass
The sfftk.formats.seg.SeggerSegmentation class’s initialiser does the reading using the sfftk.readers.segreader module described in Step I: Create a reader module;
class SeggerSegmentation(Segmentation):
def __init__(self, fn, top_level=False, *args, **kwargs):
"""Initialise the reader"""
self._fn = fn
self._segmentation = segreader.get_data(self._fn, *args, **kwargs)
self._top_level = top_level
We then reference the header and segments attributes using propers to decouple the actual hosted objects from the user-facing attributes.
class SeggerSegmentation(Segmentation):
# ...
@property
def header(self):
"""The header for this segmentation"""
return SeggerHeader(self._segmentation)
and delegate the sfftk.formats.seg.SeggerHeader to expose header attributes.
class SeggerHeader(Header):
"""Header class"""
def __init__(self, segmentation):
self._segmentation = segmentation
@property
def name(self):
"""The name of segmentation"""
return self._segmentation.format
@property
def version(self):
"""The version of Segger used"""
return self._segmentation.format_version
@property
def map_path(self):
"""The path to the original segmented map"""
return self._segmentation.map_path
@property
def ijk_to_xyz_transform(self):
"""The image-to-physical transform"""
return self._segmentation.ijk_to_xyz_transform
@property
def file_path(self):
"""The path to the .seg file"""
return self._segmentation.file_path
@property
def root_parent_ids(self):
"""Parent IDs for root segments"""
return self._segmentation.root_parent_ids
@property
def region_ids(self):
"""All region IDs"""
return self._segmentation.region_ids
@property
def parent_ids(self):
"""All parent IDs"""
return self._segmentation.parent_ids
@property
def map_size(self):
"""Map dimensions"""
return self._segmentation.map_size
@property
def mask(self):
return self._segmentation.mask
@property
def simplified_mask(self):
return self._segmentation.simplify_mask(self.mask)
Notice that all underlying functionality is obtained from the ad hoc reader, which does the heavy lifting. The adapter merely adapts the ad hoc sfftk.readers.segreader.SeggerSegmentation class for the EMDB-SFF schema.
Segments are contained in a list and exposed through the segments property of the sfftk.formats.seg.SeggerSegmentation class.
class SeggerSegmentation(Segmentation):
# ...
@property
def segments(self):
"""The segments in this segmentation"""
if self._top_level:
segments = [SeggerSegment(self._segmentation, region_id) for region_id in self.header.root_parent_ids]
else:
segments = [SeggerSegment(self._segmentation, region_id) for region_id in self.header.region_ids if
region_id != 0]
return segments
To perform a conversion the sfftk.formats.seg.SeggerSegmentation class implements a sfftk.formats.seg.SeggerSegmentation.convert() method that uses the EMDB-SFF schema API as described in Developing with sfftk.
class SeggerSegmentation(Segmentation):
# ...
def convert(self, args, *_args, **_kwargs):
"""Method to convert a :py:class:`sfftkrw.SFFSegmentation` object"""
segmentation = schema.SFFSegmentation()
segmentation.name = "Segger Segmentation"
segmentation.software = schema.SFFSoftware(
name=self.header.name,
version=self.header.version,
)
segmentation.transforms = schema.SFFTransformList()
segmentation.transforms.add_transform(
schema.SFFTransformationMatrix(
rows=3,
cols=4,
data='1.0 0.0 0.0 1.0 0.0 1.0 0.0 1.0 0.0 0.0 1.0 1.0'
)
)
segmentation.transforms.add_transform(
schema.SFFTransformationMatrix(
rows=3,
cols=4,
data=" ".join(map(str, self.header.ijk_to_xyz_transform.flatten().tolist()))
)
)
segmentation.primaryDescriptor = "threeDVolume"
segments = schema.SFFSegmentList()
for s in self.segments:
segment = s.convert()
segments.add_segment(segment)
# finally pack everything together
segmentation.segments = segments
# lattice
segmentation.lattices = schema.SFFLatticeList()
cols, rows, sections = self.header.map_size
lattice = schema.SFFLattice(
mode='uint32',
endianness='little',
size=schema.SFFVolumeStructure(cols=cols, rows=rows, sections=sections),
start=schema.SFFVolumeIndex(cols=0, rows=0, sections=0),
data=self.header.simplified_mask
)
segmentation.lattices.add_lattice(lattice)
# details
if args.details is not None:
segmentation.details = args.details
elif 'details' in _kwargs:
segmentation.details = _kwargs['details']
return segmentation
Notice that in the loop for each segment (for s in self.segments) we refer to each segment’s convert() method which is implemented as follows:
class SeggerSegment(Segment):
# ...
def convert(self, **kwargs):
"""Convert to a :py:class:`sfftkrw.SFFSegment` object"""
segment = schema.SFFSegment()
segment.id = self.region_id
segment.parentID = self.parent_id
# annotation
segment.annotation, segment.colour = self.annotation.convert()
# geometry
# segment.volume = self.volume.convert()
segment.volume = schema.SFFThreeDVolume()
segment.volume.latticeId = 0
segment.volume.value = self.region_id
return segment
and which in turn calls the convert() methods on the contained annotation and volume objects.
Step III: Implement a convert hook in the sfftk.sff.handle_convert() function
In order to perform conversion using the command-line utility, we need to have the convert utility recognise segmentations by file type. This is simply done by add adding a regular expression match object in the sfftk.sff.handle_convert() function as follows:
def handle_convert(args, configs): # @UnusedVariable
# ...
if args.multi_file:
# segmentation file formats that support multi-file segmentations will be handled here
# ...
else:
if re.match(r'.*\.mod$', args.from_file, re.IGNORECASE):
# handle reading of IMOD files
elif re.match(r'.*\.seg$', args.from_file, re.IGNORECASE):
from .formats.seg import SeggerSegmentation
seg = SeggerSegmentation(args.from_file, top_level=args.top_level_only)
# other file formats go here
else:
raise ValueError("Unknown file type %s" % args.from_file)
# export (convert first if needed)
# ...
sff_seg.export(args.output)
return os.EX_OK
Once this is done the following should work:
sff convert --top-level-only file.seg --verbose
Step IV: Add test data files to the sfftk/test_data folder
Provide an example segmentation file in the sfftk.test_data package.
Step V: Add unit tests in the sfftk.unittests module
Write unit tests and add them to the sfftk.unittests.test_formats module. Each format that you add should implement a read and convert test method (respectively called test_<format>_read and test_<format>_convert. See the sfftk.unittests.test_formats module for examples.
Of course, you are welcome to contact us for help doing the above. Also, if you have any suggestions on how to improve the extension process these are invited.
Once all this components are in place conversion to XML, HDF5 and JSON should be automatic.
sff convert file.<format> -f sff
sff convert file.<format> -f hff
sff convert file.<format> -f json
Also, tests can be run as follows:
sff tests formats
Adding A Search Resource
The list of search resources is extensible. Any new search results must fulfil the following criteria:
Have a A REST API;
Return the result as either JSON or TAB-delimitted text;
Step I: Add the resource description
The resource descriptions are contained in sfftk.notes.__init__.py. For example, the one for
OLS is:
RESOURCE_LIST['ols'] = {
'name': 'OLS',
'root_url': 'https://www.ebi.ac.uk/ols/api/',
'format': 'json',
'result_path': ['response', 'docs'],
'result_count': ['response', 'numFound'],
}
RESOURCE_LIST is a dictionary whose primary key is the lower-case abbreviation of the resource name (ols here).
Each resource’s description is a dictionary of:
name- proper abbreviation of the resource’s name (UniProtnotUniprot);root_url- the part of the REST API which is invariant i.e. excluding search parameters;format- eitherjsonortab;result_path- the part of the response data that contains a list of results. In the above example,response,docsare a path to a list element of the JSON where all results are found. Ifformatistabset this toNone.result_count- the part of the response data that contains the count of the complete results. Usually only a page of all results is returned but it is useful to inform the user of how many results were found. If this is not present set it toNone.
Step II: Add a handler to return the correct URL
Modify the sfftk.notes.find.SearchResults.get_url() method by adding a conditional handler for
the resource that returns the complete search URL (i.e. including the search term and additional options allowed on the
REST API).
The if statement uses the resource name to identify the resource.
For example, the URL for OLS is generated as follows:
# ols
if self.name == 'OLS':
url = self.root_url + "search?q={}&start={}&rows={}".format(
self.search_args.search_term,
self.search_args.start - 1,
self.search_args.rows,
)
if self.search_args.ontology:
url += "&ontology={}".format(self.search_args.ontology)
if self.search_args.exact:
url += "&exact=on"
if self.search_args.obsoletes:
url += "&obsoletes=on"
Each of the subsequent if statements conditionally include additional parameters specified in the command-line
arguments.
Step III: Define how the table is formated
The sfftk.notes.find.SearchResource.tabulate() specifies how to tabulate results from each resource. We use
two classes: sfftk.notes.find.TableField and sfftk.notes.find.ResultsTable to do this. Here is
an example for OLS:
if self._resource.name == 'OLS':
fields = [
TableField(u'index', key=u'index', pc=5, is_index=True, justify=u'right'),
TableField(u'label', key=u'label', pc=10),
TableField(u'resource', key=u'ontology_name', pc=5, justify=u'center'),
TableField(u'url', key=u'iri', pc=30),
TableField(u'accession', key=u'short_form', pc=10, justify=u'center'),
TableField(u'description', key=u'description', pc=40, is_iterable=True),
]
table += _str(ResultsTable(self, fields=fields))
You will need to refer to the resource’s REST API documentation to match result fields with table fields.
sfftk.notes.find.TableField accepts the following arguments:
The first (positional) argument is the name of the field and appears in the table header;
keyis the official name of the field in the REST API; thesfftk.notes.find.TableFielduses the key to extract the row values;Instead of using the
keyargument, the user may usetextwhich will be a fixed value for all rows;pcis the field width in percent; alternatively, use thewidthargument to specify how many characters wide the field should be;is_indexsets the field to be an index field i.e. to show the row number;justifycan either beright,leftorcenterand specifies the field text alignment;_formatspecifies a format string (a string with with{}which will replace{}with the field value. This is helpful when creating URLs/IRIs to an accession page e.g.TableField('url', key='EntryID', _format='https://www.ebi.ac.uk/pdbe/emdb/EMD-{}', pc=30),
for an EMDB entry page.
Step IV: Write tests and test
You will then need to write at least two tests in the sfftk.unittests.test_notes.TestNotesFindSearchResource class:
parser tests that check that the command-line arguments are correct (see the module for other examples);
get_url tests that ensure that the URL is correctly formed for all options;
Step IV: Write documentation for the new resource
Finally, document the resource in the section Specifying The Resource To Search by describing the following:
the name and link to the resource in the list of resources;
the keyword to be passed to the
-R/--resourceflag;