Converting Files To EMDB-SFF
Introduction
Converting files to use the EMDB-SFF data model is one of the core functions
of sfftk. This guide describes in detail how to accomplish conversions.
Synopsis
Running
sff convert
sff convert -h
sff convert --help
displays all conversion options.
sff convert
usage: sff convert [-h] [-D DETAILS] [-R PRIMARY_DESCRIPTOR] [-v] [-x]
[--json-indent JSON_INDENT] [--json-sort]
[-o OUTPUT | -f FORMAT] [-p CONFIG_PATH] [-b] [-a] [-m]
[--subtype-index SUBTYPE_INDEX] [--image IMAGE]
[--label-tree LABEL_TREE]
[--subtomogram-average SUBTOMOGRAM_AVERAGE]
[--image-name-field IMAGE_NAME_FIELD]
[--euler-angle-convention {zyz,zxz,xyx,xzx,yxy,yzy}]
[--radians]
[from_file [from_file ...]]
Perform conversions to EMDB-SFF
positional arguments:
from_file file to convert from
optional arguments:
-h, --help show this help message and exit
-D DETAILS, --details DETAILS
populates <details>...</details> in the XML file
-R PRIMARY_DESCRIPTOR, --primary-descriptor PRIMARY_DESCRIPTOR
populates the
<primary_descriptor>...</primary_descriptor> to this
value [valid values: three_d_volume, mesh_list,
shape_primitive_list]
-v, --verbose verbose output
-x, --exclude-geometry
do not include the geometry in the conversion;
geometry is included by default [default: False]
--json-indent JSON_INDENT
size in spaces of the JSON indent [default: 2]
--json-sort output JSON sorted lexicographically [default: False]
-o OUTPUT, --output OUTPUT
file to convert to; the extension (.sff, .hff, .json)
determines the output format [default: None]
-f FORMAT, --format FORMAT
output file format; valid options are: sff (XML), hff
(HDF5), json (JSON) [default: sff]
-p CONFIG_PATH, --config-path CONFIG_PATH
path to configs file
-b, --shipped-configs
use shipped configs only if config path and user
configs fail [default: False]
-a, --all-levels for segments structured hierarchically (e.g. Segger
from UCSF Chimera and Chimera X) convert all segment
leves in the hierarchy [default: False]
-m, --multi-file enables convert to treat multiple files as individual
segments of a single segmentation; only works for the
following filetypes: stl, map, mrc, rec, star
[default: False]
--subtype-index SUBTYPE_INDEX
some file extensions are used by multiple file types
--image IMAGE specify the segmented EMDB MAP/MRC file from which to
determine the correct image-to-physical transform
--label-tree LABEL_TREE
a JSON file produced by running 'sff prep mergemask'
which captures: 1) the mask labels (key:
'mask_to_label') and 2) the hierarchical relationship
between labels (key: 'label_tree')
--subtomogram-average SUBTOMOGRAM_AVERAGE
the result of subtomogram averaging or a particle mask
for visualisation in CCP4 format (.mrc, .map, .rec)
--image-name-field IMAGE_NAME_FIELD
the field in the star file that contains the image
name [default: '_rlnImageName']
--euler-angle-convention {zyz,zxz,xyx,xzx,yxy,yzy}
the Euler angle convention used in the subtomogram
averaging [default: 'zyz' - case insensitive]
--radians use radians instead of degrees for Euler angles
[default: False i.e. use degrees]
Quick Start
Output to XML (Default)
sff convert file.seg
Note
New in version 0.7.0.
Without any other arguments, the above command will print out the following warning:
Warning: missing --image <file.map> option to accurately determine image-to-physical transform
sfftk assumes that the original segmented image is specified as an MRC-like file e.g. *.map (images deposited into EMDB), *.mrc or compatible formats such as *.rec used by IMOD, which have metadata to compute this transform.
Specifying the Segmented Image File
First, ensure that the segmented image is specified in an MRC-like format. For images deposited in the EMDB use the deposited image.
Use the --image argument to specify the segmented image.
sff convert --image file.map file.seg
Some segmentation file formats specify the transform and sfftk will fall back on this if the image is not specified.
Note
Viewing transforms
To view the transform inferred from the image file use sff view --transform file.map. Visit the view documentation pages Show Image-to-Physical Transform for more information.
Specify Output File
sff convert file.seg -o file.sff
sff convert file.seg --output /path/to/output/file.sff
sff convert file.seg -o file.hff
sff convert file.seg -o --exclude-geometry file.json # only metadata; no geometrical data
Specify Output Format
sff convert file.seg -f hff
sff convert file.seg --format hff
EMDB-SFF Format Interconversion
sff convert file.sff --output /path/to/output/file.hff
sff convert file.hff --format json
sff convert file.sff --format sff # reduntant but should work
Verbose Operation
sff convert -v file.hff
sff convert --verbose file.hff
Include All Segments
When a segmentation is defined hierarchically only the top level of segments (i.e. those just under the root) will
be included by default. Use -a/--all-levels argument to include all segments. This can lead to very large files.
Segger (.seg) segmentations are one such example.
sff convert -a file.seg
sff convert --all-levels file.seg
Set Details
sff convert -D "Lorem ipsum dolor..." file.seg # strings must be quoted (single/double)
sff convert --details "Lorem ipsum dolor..." file.seg
Change Primary Descriptor
sff convert -R shape_primitive_list file.surf # IMOD file
sff convert --primary-descriptor shape_primitive_list file.surf # IMOD file
Input Formats
sfftk can convert several segmentation file formats (see
Supported Formats) into EMDB-SFF files.
Output Formats
EMDB-SFF files can be output as XML (.sff, .xml), HDF5 (.hff, .h5 or .hdf5) or JSON
(.json).
Both XML and HDF5 are quite compact and in many cases would be smaller than the original segmentation file.
JSON EMDB-SFF files may exclude geometry if created with
-x/--exclude-geometryflag; they are primarily used as temporary files during annotation for speed.Interconversion of the three formats is lossless (with the exception of geometrical data when converting to JSON - if geometrical data is excluded).
There are two ways to perform conversion:
Specifying the output path with
-o/--outputflagSpecifying the output format with
-f/--formatflag
Specifying the output path with -o/--output flag
Conversion is performed as follows:
sff convert file.seg -o file.sff
sff convert file.seg --output /path/to/output/file.sff
The output file extension determines the output format i.e.
sff convert file.seg -o file.hff
will result in an HDF5 file while
sff convert file.seg --output file.json
will be a JSON file.
Specifying the output format with -f/--format flag
The -f/–format options ensures that the output file will be in the same
directory as the original segmentation file. The -f flag takes one of three
values:
sfffor XML fileshfffor HDF5 filesjsonfor JSON files.
Any other value raises an error.
sff convert file.seg -f hff
sff convert file.seg --format hff
The default format (if none is specified) is sff (XML).
sff convert file.seg
results in file.sff as output.
EMDB-SFF Format Interconversion
It is also possible to perform interconversions between XML, HDF5 and JSON EMDB-SFF files.
sff convert file.sff --output /path/to/output/file.hff
or using --format
sff convert file.hff --format json
Even null conversions are possible:
sff convert file.sff --format sff
Conversions from JSON to XML/HDF5 where the latter excluded the geometry will not reinstate the geometrical description information.
Verbose Operation
As with many Linux shell programs the -v/--verbose option prints status
information on the terminal.
sff convert --verbose file.hff
Tue Sep 12 15:29:18 2017 Seting output file to file.sff
Tue Sep 12 15:29:18 2017 Converting from EMDB-SFF (HDF5) file file.hff
Tue Sep 12 15:30:03 2017 Created SFFSegmentation object
Tue Sep 12 15:30:03 2017 Exporting to file.sff
Tue Sep 12 15:30:07 2017 Done
Including All Segments
Segger segmentations include hundreds to thousands of sub-segmentations due to how the algorithm it uses (watershed algorithm) to segment the volume. The segmentations thus form a tree with the root having an ID of zero. Mostly, we are only interested in the children of the root which are in themselves roots of another tree. By default we only transfer the children of the global root into the EMDB-SFF file.
Consider the following tree of segments:
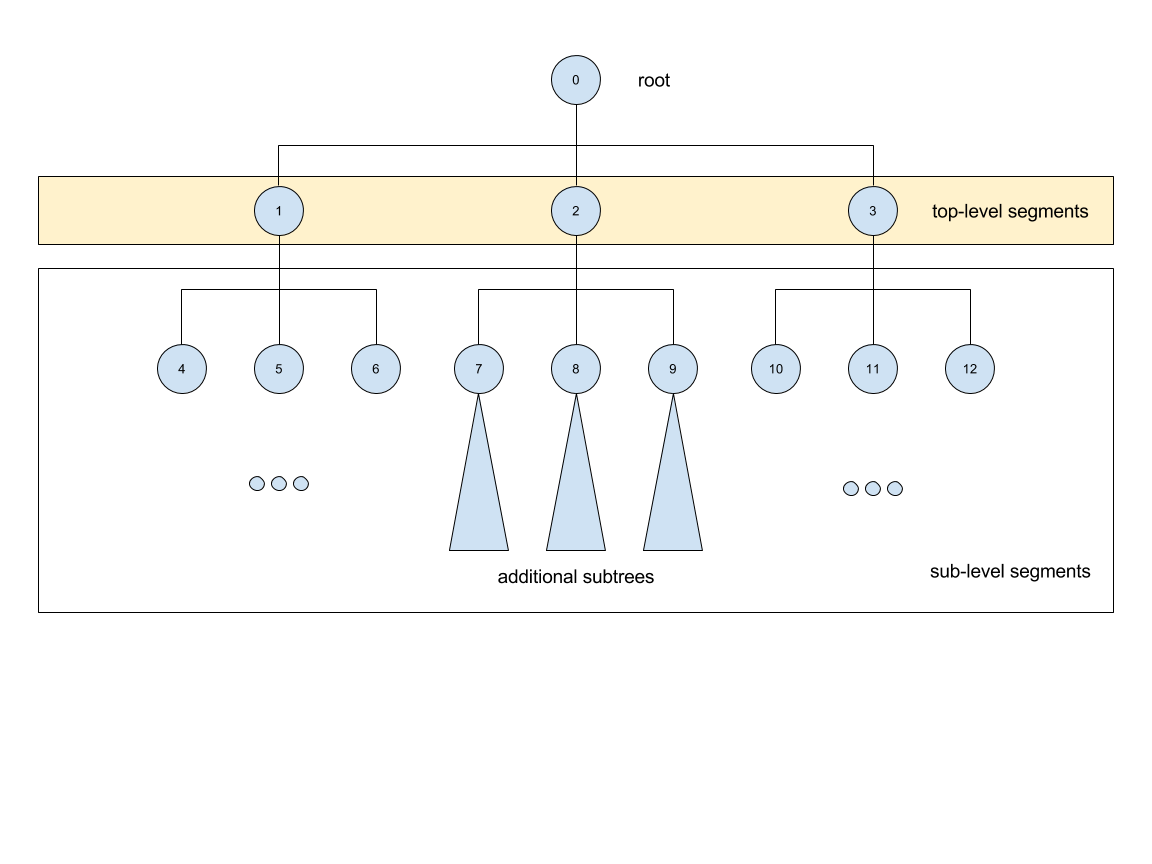
The segmentation contains different levels commencing from the root down, with
children segments contained within parent segments. Specifying
-a/--all-levels treats all children of the root as segments and
includes all segments. Therefore, running
sff convert --all-levels file.seg
on the above will produce an EMDB-SFF file with hundreds of segments. The default operation results in a more compact file.
Specify Details
The EMDB-SFF data model provides for an optional <details/> tag for
auxilliary information. The contents of this option will be put into
<details/>.
sff convert --details "Lorem ipsum dolor..." file.seg
Todo
Allow a user to pass a file whose contents will be inserted into <details/>.
Changing The Primary Descriptor
The EMDB-SFF data model provides for three possible geometrical descriptors: meshes (mesh_list), shape primitives (shape_primitive_list) and 3D volumes (three_d_volume).
In some cases, such as with IMOD segmentations, more than one geometrical descriptor may have been specified for the same segmentations.
The mandatory <primaryDescriptor/> field specifies the main geometrical
descriptor to be used when performing conversions and other processing tasks.
Only valid values are allowed; otherwise a ValueError is raised.
The table below shows valid primary descriptors by file type.
File format |
Valid primary descriptors |
|---|---|
AmiraMesh |
three_d_volume |
AmiraHxSurface |
mesh_list |
SuRVoS |
three_d_volume |
CCP4 masks |
three_d_volume |
IMOD |
mesh_list (default), shape_primitive_list |
Segger |
three_d_volume |
STL |
mesh_list |
Note
IMOD files must have a mesh generated using imodmesh command. Open contours will need to be converted to
tubes using the -t <obj_list> option. For example, for an IMOD file file.mod with three objects all of
which are open contours we can run:
~$ imodmesh -t 1,2,3 -d 10 -E -Z 1.0 file.mod
which convert to tubes objects 1, 2 and 3 (-t 1,2,3), cap ends (-E) with domes at a scale of 1.0
(-Z 1.0) and a diameter of 10 pixels (-d 10).
You can find out much more about using imodmesh at its documentation page.
Note that the primary descriptor should only be changed to a value of a geometrical descriptor that is actually present in the EMDB-SFF file.
Working with Multifile Segmentations
Some of the segmentation file formats supported are designed to hold one segment per file. Therefore, representing a complete segmentation will require multiple files.
Currently the following file formats are multifile by design:
CCP4 and related files - these files store segments as a 3D volume with segment region marked by specific voxel values (e.g.
1for in segment voxels and0for the background. Specific file formats have.mrc,.mapand.rec.Stereolithography files - while it is possible to concatenate several STL files into one, STL files do not contain metadata such as segment colour. Therefore, it is best to handle them as multifiles. STL files have a
.stlextension.
Multifiles utilise the -m/--multi-file argument followed by all the files each of which should
specify a single segment.
sff convert -m file1.map file2.map file3.map
The above command will use default options and write an EMDB-SFF file to file1.sff. Alternatively,
the user should specify the output file
sff convert -m file1.map file2.map file3.map --output file.sff
Converting Subtomogram Averages
Subtomogram averages are the result of subtomogram averaging and are typically stored in CCP4 format
(.mrc, .map, .rec). sfftk currently only works with RELION subtomogram averages in .star format.
Conversion of subtomogram averages is similar to that of other file formats in that
the user specifies the file to convert and the output file. Optionally, the user may provide the image file from which
the subtomogram average was derived using the --image option to accurately determine the image-to-physical transform.
sff convert --image file.map file.star
Specifying the Particle File
It is also possible to specify the particle file from which the subtomogram average was derived using the --subtomogram-average option. This is useful when the subtomogram average is a particle mask for visualisation. However, given that the particle will be of higher resolution that the original tonmogram, the user may need to downsample the particle for visualisation purposes.
sff convert --subtomogram-average particle.mrc --image image.mrc file.star
A tool that can be used to prepare artificial particles is available from https://github.com/emdb-empiar/masks.git.
Multiple Subtomogram Averages
As with multifile segmentations, it is possible to convert multiple subtomogram averages into a single EMDB-SFF file. The user should use the -m/--multi-file option to specify the subtomogram averages. However, only one --subtomogram-average option should be used to specify the particle file.
sff convert -m file1.star file2.star file3.star --output file.sff
Specifying Euler Angle Convention
The Euler angle convention used in the subtomogram averaging can be specified using the --euler-angle-convention option. The default is zyz. The case is not important (zyz``=``ZYZ).
Specifying Angle Type
The user can specify whether the Euler angles are in degrees or radians using the --radians option. The default is degrees.
Specifying Configurations To Use
sfftk makes use of persistent configurations which affect how certain operations
are performed. There are three types of configurations detailed in the dedicated
documentation on configs (see Where Configurations Are Stored) in decreasing order of priority:
custom configs defined in a
path/to/sff.conffile;user configs stored in
~/.sfftk/sff.conf;shipped configs which will sit with the installed
sfftkpackage.
Custom configs are invoked using the -p/--config-path option:
sff convert -p path/to/configs file.seg
sff convert --config-path path/to/configs file.seg
User configs are default and require no special flags.
Shipped configs use the -b/--shipped-configs flag with no arguments:
sff convert -b file.am
sff convert --shipped-configs file.am